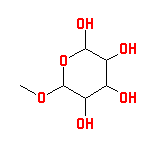
|
| A structure drawing created with the Java Molecular Editor available at the Corina web site. |
In favorable cases, atomic coordinates for a given hetero compound, or at least for a similar compound, can be found in the PDB. However, instead of looking in the PDB directly, often it is much more convenient to search for hetero compounds on Gerard Kleywegt's HIC-UP server.
The most comprehensive resource for small molecules structures is the Cambridge Structural Database (CSD). At the time of this writing, the CSD contains crystal structure information for over 190,000 organic and metal organic compounds. The coordinates are in general of high quality and very well suited for the derivation of topology and parameter definitions. However, the CSD is a commercial product and therefore its use is limited to members of registered, paying institutions.
The next best resource for obtaining coordinates, or even ready-to-use CNS topology and parameter files, are the BIOSCI news groups. If the compound is not too exotic, there is a fair chance that someone responds to a request for hetero compound coordinates.
If all else fails, one can use commercial software or the (essentially) free web-based tool Corina to build a compound from scratch and get the conformation by energy minimization. Corina provides a tool for drawing a structure, performs minimization and returns the coordinates in PDB format.

|
| A structure drawing created with the Java Molecular Editor available at the Corina web site. |
The problem with coordinates built from scratch and with energy minimization is that the resulting conformation frequently is not the one observed in the crystal structure. In case of doubt, the best approach is to built the molecule based on the experimental electron density, optionally followed by energy minimization.
Remarks /var/tmp/xdict_8678.top
Remarks Created by XPLO2D V. 990106/2.8.4 at Fri Jul 2 03:17:51 1999 for root
Remarks Auto-generated by XPLO2D from file /var/tmp/xdict_8678.pdb
Remarks You *MUST* check/edit MASSes and CHARges !!!
Remarks Check DONOrs and ACCEptors
Remarks Verify IMPRopers yourself
Remarks DIHEdrals which are not flat are commented out
set echo=false end
{ edit masses if necessary }
MASS CX1 13.01900 ! assuming C -> 12.01100 + 1.008 * 1 (Hs)
MASS CX2 13.01900 ! assuming C -> 12.01100 + 1.008 * 1 (Hs)
MASS CX3 13.01900 ! assuming C -> 12.01100 + 1.008 * 1 (Hs)
MASS CX4 13.01900 ! assuming C -> 12.01100 + 1.008 * 1 (Hs)
MASS CX5 13.01900 ! assuming C -> 12.01100 + 1.008 * 1 (Hs)
MASS CX6 14.02700 ! assuming C -> 12.01100 + 1.008 * 2 (Hs)
MASS CX7 15.03500 ! assuming C -> 12.01100 + 1.008 * 3 (Hs)
MASS OX8 15.99900 ! assuming O -> 15.99900 + 1.008 * 0 (Hs)
MASS OX9 17.00700 ! assuming O -> 15.99900 + 1.008 * 1 (Hs)
MASS OX10 17.00700 ! assuming O -> 15.99900 + 1.008 * 1 (Hs)
MASS OX11 17.00700 ! assuming O -> 15.99900 + 1.008 * 1 (Hs)
MASS OX12 15.99900 ! assuming O -> 15.99900 + 1.008 * 0 (Hs)
MASS OX13 17.00700 ! assuming O -> 15.99900 + 1.008 * 1 (Hs)
autogenerate angles=true end
RESIdue MMA
GROUp
ATOM C1 TYPE CX1 CHARge 0.0 END ! Nr of Hs = 1
ATOM C2 TYPE CX2 CHARge 0.0 END ! Nr of Hs = 1
ATOM C3 TYPE CX3 CHARge 0.0 END ! Nr of Hs = 1
ATOM C4 TYPE CX4 CHARge 0.0 END ! Nr of Hs = 1
ATOM C5 TYPE CX5 CHARge 0.0 END ! Nr of Hs = 1
ATOM C6 TYPE CX6 CHARge 0.0 END ! Nr of Hs = 2
ATOM C7 TYPE CX7 CHARge 0.0 END ! Nr of Hs = 3
ATOM O1 TYPE OX8 CHARge 0.0 END ! Nr of Hs = 0
ATOM O2 TYPE OX9 CHARge 0.0 END ! Nr of Hs = 1
ATOM O3 TYPE OX10 CHARge 0.0 END ! Nr of Hs = 1
ATOM O4 TYPE OX11 CHARge 0.0 END ! Nr of Hs = 1
ATOM O5 TYPE OX12 CHARge 0.0 END ! Nr of Hs = 0
ATOM O6 TYPE OX13 CHARge 0.0 END ! Nr of Hs = 1
BOND C1 C2 BOND C1 O1 BOND C1 O5 BOND C2 C3
BOND C2 O2 BOND C3 C4 BOND C3 O3 BOND C4 C5
BOND C4 O4 BOND C5 C6 BOND C5 O5 BOND C6 O6
BOND C7 O1
{ edit these DIHEdrals if necessary }
! DIHEdral O1 C1 C2 C3 ! flexible dihedral ??? -68.44
DIHEdral O1 C1 C2 O2 ! flat ? (180 degrees = trans) 172.04
! DIHEdral O5 C1 C2 C3 ! flexible dihedral ??? 52.40
! DIHEdral O5 C1 C2 O2 ! flexible dihedral ??? -67.11
! DIHEdral C2 C1 O5 C5 ! flexible dihedral ??? -59.84
! DIHEdral O1 C1 O5 C5 ! flexible dihedral ??? 58.61
! DIHEdral C1 C2 C3 C4 ! flexible dihedral ??? -51.56
DIHEdral C1 C2 C3 O3 ! flat ? (180 degrees = trans) 188.46
! DIHEdral O2 C2 C3 C4 ! flexible dihedral ??? 67.39
! DIHEdral O2 C2 C3 O3 ! flexible dihedral ??? -52.59
! DIHEdral C2 C3 C4 C5 ! flexible dihedral ??? 51.91
DIHEdral C2 C3 C4 O4 ! flat ? (180 degrees = trans) 173.78
DIHEdral O3 C3 C4 C5 ! flat ? (180 degrees = trans) 171.20
! DIHEdral O3 C3 C4 O4 ! flexible dihedral ??? -66.93
DIHEdral C3 C4 C5 C6 ! flat ? (180 degrees = trans) 185.69
! DIHEdral C3 C4 C5 O5 ! flexible dihedral ??? -54.04
! DIHEdral O4 C4 C5 C6 ! flexible dihedral ??? 63.72
DIHEdral O4 C4 C5 O5 ! flat ? (180 degrees = trans) 183.98
! DIHEdral C4 C5 C6 O6 ! flexible dihedral ??? 57.85
! DIHEdral O5 C5 C6 O6 ! flexible dihedral ??? -63.23
! DIHEdral C4 C5 O5 C1 ! flexible dihedral ??? 60.78
DIHEdral C6 C5 O5 C1 ! flat ? (180 degrees = trans) 186.34
{ edit these IMPRopers if necessary }
IMPRoper C1 C2 O1 O5 ! chirality or flatness improper -34.29
IMPRoper C2 C1 C3 O2 ! chirality or flatness improper 35.51
IMPRoper C3 C2 C4 O3 ! chirality or flatness improper 35.27
IMPRoper C4 C3 C5 O4 ! chirality or flatness improper -32.96
IMPRoper C5 C4 C6 O5 ! chirality or flatness improper -35.51
{ edit any DONOrs and ACCEptors if necessary }
ACCEptor O1 C1
ACCEptor O2 C2
ACCEptor O3 C3
ACCEptor O4 C4
ACCEptor O5 C1
ACCEptor O6 C6
END { RESIdue MMA }
Remarks /var/tmp/xdict_8678.par
Remarks Created by XPLO2D V. 990106/2.8.4 at Fri Jul 2 06:12:27 1999 for root
Remarks Auto-generated by XPLO2D from file /var/tmp/xdict_8678.pdb
Remarks Parameters for residue type MMA
set echo=false end
{ edit if necessary }
BOND CX1 CX2 1000.0 1.507 ! Nobs = 1
BOND CX1 OX8 1000.0 1.416 ! Nobs = 1
BOND CX1 OX12 1000.0 1.431 ! Nobs = 1
BOND CX2 CX3 1000.0 1.523 ! Nobs = 1
BOND CX2 OX9 1000.0 1.414 ! Nobs = 1
BOND CX3 CX4 1000.0 1.537 ! Nobs = 1
BOND CX3 OX10 1000.0 1.441 ! Nobs = 1
BOND CX4 CX5 1000.0 1.539 ! Nobs = 1
BOND CX4 OX11 1000.0 1.418 ! Nobs = 1
BOND CX5 CX6 1000.0 1.534 ! Nobs = 1
BOND CX5 OX12 1000.0 1.423 ! Nobs = 1
BOND CX6 OX13 1000.0 1.429 ! Nobs = 1
BOND CX7 OX8 1000.0 1.435 ! Nobs = 1
{ edit if necessary }
ANGLe CX2 CX1 OX8 500.0 107.44 ! Nobs = 1
ANGLe CX2 CX1 OX12 500.0 109.20 ! Nobs = 1
ANGLe OX8 CX1 OX12 500.0 111.31 ! Nobs = 1
ANGLe CX1 CX2 CX3 500.0 111.76 ! Nobs = 1
ANGLe CX1 CX2 OX9 500.0 107.84 ! Nobs = 1
ANGLe CX3 CX2 OX9 500.0 108.78 ! Nobs = 1
ANGLe CX2 CX3 CX4 500.0 111.47 ! Nobs = 1
ANGLe CX2 CX3 OX10 500.0 108.03 ! Nobs = 1
ANGLe CX4 CX3 OX10 500.0 109.20 ! Nobs = 1
ANGLe CX3 CX4 CX5 500.0 110.01 ! Nobs = 1
ANGLe CX3 CX4 OX11 500.0 110.40 ! Nobs = 1
ANGLe CX5 CX4 OX11 500.0 110.23 ! Nobs = 1
ANGLe CX4 CX5 CX6 500.0 115.75 ! Nobs = 1
ANGLe CX4 CX5 OX12 500.0 108.52 ! Nobs = 1
ANGLe CX6 CX5 OX12 500.0 107.00 ! Nobs = 1
ANGLe CX5 CX6 OX13 500.0 109.50 ! Nobs = 1
ANGLe CX1 OX8 CX7 500.0 118.00 ! Nobs = 1
ANGLe CX1 OX12 CX5 500.0 117.07 ! Nobs = 1
{ edit if necessary }
DIHEdral OX8 CX1 CX2 CX3 750.0 0 -60.00 ! Nobs = 1 ... Value = -68.44
DIHEdral OX8 CX1 CX2 OX9 750.0 0 180.00 ! Nobs = 1 ... Value = 172.04
DIHEdral OX12 CX1 CX2 CX3 750.0 0 60.00 ! Nobs = 1 ... Value = 52.40
DIHEdral OX12 CX1 CX2 OX9 750.0 0 -60.00 ! Nobs = 1 ... Value = -67.11
DIHEdral CX2 CX1 OX12 CX5 750.0 0 -60.00 ! Nobs = 1 ... Value = -59.84
DIHEdral OX8 CX1 OX12 CX5 750.0 0 60.00 ! Nobs = 1 ... Value = 58.61
DIHEdral CX1 CX2 CX3 CX4 750.0 0 -60.00 ! Nobs = 1 ... Value = -51.56
DIHEdral CX1 CX2 CX3 OX10 750.0 0 180.00 ! Nobs = 1 ... Value = -171.54
DIHEdral OX9 CX2 CX3 CX4 750.0 0 60.00 ! Nobs = 1 ... Value = 67.39
DIHEdral OX9 CX2 CX3 OX10 750.0 0 -60.00 ! Nobs = 1 ... Value = -52.59
DIHEdral CX2 CX3 CX4 CX5 750.0 0 60.00 ! Nobs = 1 ... Value = 51.91
DIHEdral CX2 CX3 CX4 OX11 750.0 0 180.00 ! Nobs = 1 ... Value = 173.78
DIHEdral OX10 CX3 CX4 CX5 750.0 0 180.00 ! Nobs = 1 ... Value = 171.20
DIHEdral OX10 CX3 CX4 OX11 750.0 0 -60.00 ! Nobs = 1 ... Value = -66.93
DIHEdral CX3 CX4 CX5 CX6 750.0 0 180.00 ! Nobs = 1 ... Value = -174.31
DIHEdral CX3 CX4 CX5 OX12 750.0 0 -60.00 ! Nobs = 1 ... Value = -54.04
DIHEdral OX11 CX4 CX5 CX6 750.0 0 60.00 ! Nobs = 1 ... Value = 63.72
DIHEdral OX11 CX4 CX5 OX12 750.0 0 180.00 ! Nobs = 1 ... Value = -176.02
DIHEdral CX4 CX5 CX6 OX13 750.0 0 60.00 ! Nobs = 1 ... Value = 57.85
DIHEdral OX12 CX5 CX6 OX13 750.0 0 -60.00 ! Nobs = 1 ... Value = -63.23
DIHEdral CX4 CX5 OX12 CX1 750.0 0 60.00 ! Nobs = 1 ... Value = 60.78
DIHEdral CX6 CX5 OX12 CX1 750.0 0 180.00 ! Nobs = 1 ... Value = -173.66
{ edit if necessary }
IMPRoper CX1 CX2 OX8 OX12 750.0 0 -35.000 ! Nobs = 1 ... Value = -34.293
IMPRoper CX2 CX1 CX3 OX9 750.0 0 35.000 ! Nobs = 1 ... Value = 35.510
IMPRoper CX3 CX2 CX4 OX10 750.0 0 35.000 ! Nobs = 1 ... Value = 35.268
IMPRoper CX4 CX3 CX5 OX11 750.0 0 -35.000 ! Nobs = 1 ... Value = -32.958
IMPRoper CX5 CX4 CX6 OX12 750.0 0 -35.000 ! Nobs = 1 ... Value = -35.506
{ edit if necessary }
NONBonded CX1 0.1200 3.7418 0.1000 3.3854 ! assuming Carbon
NONBonded CX2 0.1200 3.7418 0.1000 3.3854 ! assuming Carbon
NONBonded CX3 0.1200 3.7418 0.1000 3.3854 ! assuming Carbon
NONBonded CX4 0.1200 3.7418 0.1000 3.3854 ! assuming Carbon
NONBonded CX5 0.1200 3.7418 0.1000 3.3854 ! assuming Carbon
NONBonded CX6 0.1200 3.7418 0.1000 3.3854 ! assuming Carbon
NONBonded CX7 0.1200 3.7418 0.1000 3.3854 ! assuming Carbon
NONBonded OX8 0.1591 2.8509 0.1591 2.8509 ! assuming Oxygen
NONBonded OX9 0.1591 2.8509 0.1591 2.8509 ! assuming Oxygen
NONBonded OX10 0.1591 2.8509 0.1591 2.8509 ! assuming Oxygen
NONBonded OX11 0.1591 2.8509 0.1591 2.8509 ! assuming Oxygen
NONBonded OX12 0.1591 2.8509 0.1591 2.8509 ! assuming Oxygen
NONBonded OX13 0.1591 2.8509 0.1591 2.8509 ! assuming Oxygen
set echo=true end