The search model represents only approximately half of the contents of the asymmetric unit. However, the location of one monomer is already known (from the previous rotation/translation search). This information can be used in the second translation search. The location of the first molecule will remain fixed while the location of second is tested for each rotation peak. Before this can be done, the structural information from the 2 models must be merged into single PDB and MTF files. This is performed with the CNS task file merge_structures.inp. The coordinates for the top translation search solution are used for the first molecule, while the starting model is used for the second molecule. This is because the first molecule will remain fixed in the subsequent translation search. It is also useful to give different segids for each molecule (in this case A and B). The use of a single letter makes transfer between CNS and programs such as O much easier.
cns_solve < merge_structures.inp > merge_structures.out [15 seconds]
Once the structures have been merged the final translation search can be performed. In the CNS task file it is possible to select those atoms which are to be used in structure factor calculation and also those atoms which are to remain fixed during the translation search:
{* select atoms to be used in the calculation of structure factors *}
{* this must include the molecule whose translation is unknown and
any molecules whose translation is already known which will remain
stationary during the translation search *}
{===>} atom_select=(known and (segid A or segid B));
{* select atoms of molecule(s) whose translation is already known *}
{* these atoms will not be translated during the translation search,
but their position may be optimized during the final rigid body
refinement *}
{===>} atom_placed=(segid A);
Before and after the translation search the rotational orientation of the search model is optimized by Patterson-Correlation (PC) refinement.
cns_solve < translation_dimer.inp > translation_dimer.out [4.5 minutes]
The translation search shows one solution with a significantly higher correlation coefficient than the other solutions (solution 8):
theta1 theta2 theta3 transX transY transZ monitor packing
R# 1 63.92 78.87 77.70 19.82 27.97 36.24 0.263 0.5974
R# 2 63.55 54.56 49.19 4.00 35.73 29.67 0.253 0.5554
R# 3 71.96 64.38 30.20 -13.82 23.56 36.53 0.287 0.5954
R# 4 224.12 88.55 206.56 16.52 11.19 24.63 0.275 0.6440
R# 5 64.05 46.94 85.10 -19.42 -14.14 38.44 0.286 0.4691
R# 6 52.96 81.20 325.88 -29.17 -32.98 41.83 0.289 0.5105
R# 7 41.77 68.31 48.33 -24.28 12.37 20.49 0.249 0.4805
R# 8 43.30 6.63 301.60 -20.31 -23.52 25.99 0.509 0.6762
R# 9 53.54 72.98 313.94 -23.47 -11.39 5.48 0.310 0.5959
R# 10 54.47 84.63 46.36 6.48 23.18 38.03 0.330 0.4735
It is also important to note that the correlation coefficient increased signficantly compared to the first translation search with only one molecule. The packing value is at a maxmimum indicating minimimal overlap for this solution compared to the others.
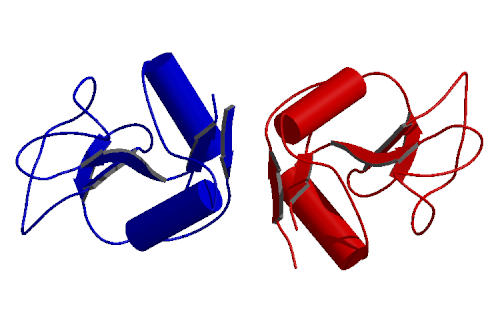
Visual inspection of the best solution shows that the two molecules are separated in space. It is best to automatically relocate the molecules so they are close together in space (making graphics work easier). This can be done by applying crystallograpic symmetry and unit cell translations in order to maximize the surface area buried between the two molecules. This is performed with the CNS task file shift_molecules.inp:
cns_solve < shift_molecules.inp > shift_molecules.out [25 seconds]
After repositioning of the molecules a compact dimer can be seen:
 |
| Complete MBP molecular replacement solution |