cns_solve < flip_sites.inp > flip_sites.out [< 1 second]
The task files for the two independent SAD phasing jobs are identical
except for the definitions of the file names for the heavy atom sites
and the output file names.
cns_solve < sad_phase2.inp > sad_phase2.out [14 minutes]
cns_solve < sad_phase2_flip.inp > sad_phase2_flip.out [14 minutes]
For the density modification, it is necessary to have a good estimate
of the solvent content. In a preliminary step, the solvent content is
estimated by using matthews_coef.inp.
cns_solve < matthews_coef.inp > matthews_coef.out [< 1 second]
The estimated solvent content shown in matthews_coef.list is between 0.53 and 0.55. density_modify.inp and density_modify_flip.inp are modified accordingly by setting the solvent content parameter to 0.54.
cns_solve < density_modify.inp > density_modify.out [27 minutes]
cns_solve < density_modify_flip.inp > density_modify_flip.out [28 minutes]
The resulting output files are:
density_modify*.list - listing file with statistics for
each modification cycle
density_modify*.hkl - reflection file with new Hendrickson-Lattman
coefficients
density_modify*.map - map file with modified electron density
density_modify*.mask - file with definition of solvent mask
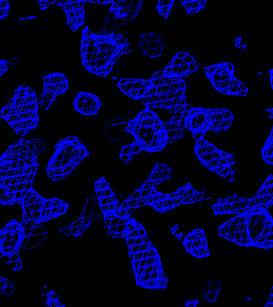
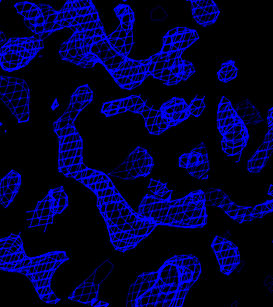
Electron density maps before solvent flipping are computed
with modified copies of the fourier_map.inp task file.
cns_solve < fourier_map_sad2.inp > fourier_map_sad2.out [21 seconds]
cns_solve < fourier_map_sad2_flip.inp > fourier_map_sad2_flip.out [21 seconds]
If you have mapman installed, you can use the command
map_to_omap *.map
to convert the CNS maps to a format which can be read into O.
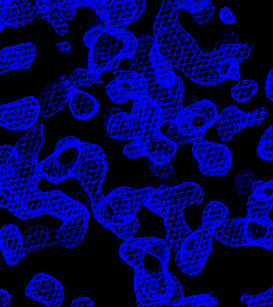
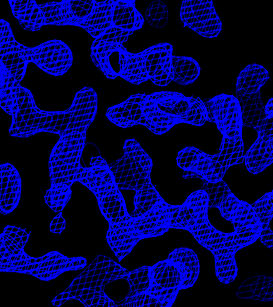
In O, enter @omac to read in the four maps. Portions
of these maps are shown below.
| Original sites | Flipped sites |
 |
 |
| Original sites | Flipped sites |
 |
 |