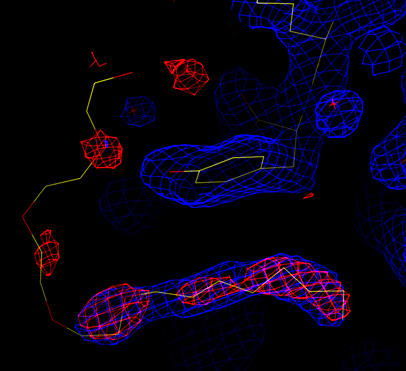
Gradient map in red (3 sigma).
The coordinates for the wild-type structure (1prn) are shown.
Density is seen for a lipid/detergent molecule (also seen in 7prn).
The model statistics are checked (as at the start of refinement) in order to check the model geometry and fit to the experimental diffraction data. In this example the CNS task file model_stats_twin_final.inp is used to analyse the model geometry and diffraction statistics:
cns_solve < model_stats_twin_final.inp > model_stats_twin_final.out [2 minutes]
The output listing file (model_stats_twin_final.list) contains a variety of information which is self-explanatory. Important things to note are:
R-values:
=================================== summary ================================== resolution range: 500.0 - 2.25 A Twinned R-values: initial r= 0.2866 free_r= 0.2467 after B-factor and/or bulk solvent correction r= 0.1425 free_r= 0.1847
R-values versus resolution:
============================= twinned R-values ===============================
=======> R-values with |Fobs|/sigma cutoff= 0.0
Test set (test = 1):
#bin | resolution range | #refl |
1 4.85 500.01 196 0.1743
2 3.85 4.85 187 0.1611
3 3.36 3.85 217 0.1649
4 3.05 3.36 211 0.1822
5 2.83 3.05 223 0.1838
6 2.67 2.83 239 0.2054
7 2.53 2.67 233 0.2191
8 2.42 2.53 215 0.2189
9 2.33 2.42 242 0.2103
10 2.25 2.33 232 0.2169
Working set:
#bin | resolution range | #refl |
1 4.85 500.01 2128 0.1347
2 3.85 4.85 2179 0.1118
3 3.36 3.85 2188 0.1148
4 3.05 3.36 2188 0.1320
5 2.83 3.05 2153 0.1562
6 2.67 2.83 2192 0.1667
7 2.53 2.67 2132 0.1790
8 2.42 2.53 2179 0.1914
9 2.33 2.42 2182 0.2014
10 2.25 2.33 2174 0.1943
This distribution of R-values is reasonable - there is no dramatic increase in R-value (in particular free R-value) as resolution increases. If there were resolution shells with R-values significantly higher than the rest this might indicate possible problems with the data processing (ice rings for example). However, the R-values indicate the fit to the experimental data and should not be manipulated by removing data - in particular by the use of sigma cutoffs to exclude weak data.
The overall geometry:
rmsd bonds= 0.008428 with 2 bond violations > 0.05 rmsd angles= 1.37251 with 9 angle violations > 8.0 rmsd dihedrals= 27.05995 with 0 angle violations > 60.0 rmsd improper= 0.65454 with 3 angle violations > 3.0
These overall statistics look very good. It is worth checking the outlying angle and improper deviations in the detailed geometry analysis (below).
The geometry in detail:
================================= geometry =================================== =======> bond violations (atom-i |atom-j ) dist. equil. delta energy const. (A 1 CG |A 1 SD ) 1.858 1.803 0.055 1.533 512.111 (A 1 SD |A 1 CE ) 1.871 1.791 0.080 1.093 170.066 =======> angle violations (atom-i |atom-j |atom-k ) angle equil. delta energy const. (A 8 N |A 8 CA |A 8 C ) 103.275 112.500 -9.225 5.991 231.085 (A 86 N |A 86 CA |A 86 C ) 102.880 111.200 -8.320 5.227 247.886 (A 91 N |A 91 CA |A 91 C ) 121.715 111.200 10.515 8.349 247.886 (A 196 N |A 196 CA |A 196 C ) 101.624 111.200 -9.576 6.925 247.886 (A 201 N |A 201 CA |A 201 C ) 102.670 111.200 -8.530 5.495 247.886 (A 214 N |A 214 CA |A 214 C ) 98.559 111.200 -12.641 12.066 247.886 (A 215 N |A 215 CA |A 215 C ) 120.040 111.200 8.840 5.901 247.886 (A 241 N |A 241 CA |A 241 C ) 100.631 111.200 -10.569 8.435 247.886 (A 271 N |A 271 CA |A 271 C ) 101.996 111.200 -9.204 6.397 247.886 =======> improper angle violations (atom-i |atom-j |atom-k |atom-L ) angle equil. delta energy const. period (A 55 CG |A 55 CD1 |A 55 CD2 |A 55 CB ) 3.244 0.000 -3.244 2.404 750.000 0 (A 102 CA |A 102 N |A 102 C |A 102 CB ) 32.058 35.264 3.206 2.349 750.000 0 (A 259 CG |A 259 CD1 |A 259 CD2 |A 259 CB ) 3.194 0.000 -3.194 2.331 750.000 0 =======> dihedral angle violations (atom-i |atom-j |atom-k |atom-L ) angle equil. delta energy const. period
Specific problems with the model geometry can be identified here. The angle deviation all occur at N-CA-C angles. Based on high resolution structures this angle is flexible, so small deviations for some of these angles are tolerated.
Non-trans peptide bonds:
============================ non-trans peptides ============================== there are no distorted or cis- peptide planes
The presence of non-trans peptides, unless they are proline residues, will cause problems in refinement. They can be identified, and an appropriate parameter file created using the CNS task file general/cis_peptide.inp. The parameter file generated is read into subsequent refinement task files.
Statistics about detwinning of the data:
================================ detwinning ==================================
data detwinned with: F_detwin = (Fo^2 - alpha*[Fo^2 + Fo'^2])/(1 - 2*alpha)
Fo'[h,k,l] = Fo[h,-h-k,-l]
alpha = 0.304
reflections rejected (I_detwin <= 0): 683
working set: 626
test set: 57
The detwinning algorithm depends on whether the twinning is perfect or partial. Some reflections are rejected during the detwinning procedure because the resultant intensity is less than zero.
Detwinned R-values:
============================ detwinned R-values ============================== resolution range: 500.0 - 2.25 A R-values: after detwinning r= 0.2060 free_r= 0.2523
The R-values after detwinning are in greater in magnitude than the twinned R-values.
After analysis of the geometry and fit to the experimental data electron density maps should be checked. Cross-validated, sigma-A weighted 2Fo-Fc and gradient maps are calculated:
cns_solve < model_map_twin_final_grad.inp > model_map_twin_final_grad.out [1 minute]
cns_solve < model_map_twin_final_2fofc.inp > model_map_twin_final_2fofc.out [1 minute]
If you have mapman installed, you can use the command
map_to_omap *.map
to convert the CNS maps to a format which can be read into
O. In O, enter @omac to read in the current
model and map.
Analysis of the gradient map and cross-validated sigma-A weighted 2Fo-Fc map shows that an ordered water molecule is still missing (not shown). Also, there is electron density in the regions where a lipid/detergent molecule was identified in the original structure refined without twinning (7prn), and a lipid/detergent molecule seen in the wild-type structure (1prn):
 |
| Cross-validated Sigma-A weighted 2Fo-Fc map in blue (at 1.5 sigma). Gradient map in red (3 sigma). The coordinates for the wild-type structure (1prn) are shown. Density is seen for a lipid/detergent molecule (also seen in 7prn). |
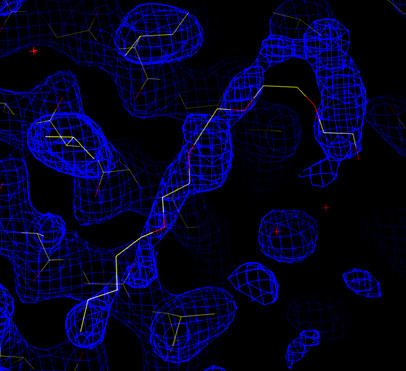 |
| Cross-validated Sigma-A weighted 2Fo-Fc map in blue (at 1.0 sigma). The coordinates for the wild-type structure (1prn) are shown. Putative density for a lipid/detergent molecule. |