
A fortran program to determine the secondary and first level supersecondary structure of a protein from alpha carbon atom coordinates.

The above is a portion of the graphical output from DEFINE_S for the protein ribonuclease A. The individual icons are described below.
DEFINE_S was written by F.M. Richards and C.E.Kundrot. It produces a list of the secondary structure of a protein and some relations between the secondary elements based solely on the coordinates of the alpha carbon atoms. The program will also, upon request, print a distance matrix of the CA atoms.
A reference describing the method is:
F.M. Richards and C.E. Kundrot (1988) Identification of Structural Motifs From Protein Coordinate Data: Secondary Structure and First-Level Supersecondary Structure. PROTEINS: Structure, Function, and Genetics 3:71-84.
To use DEFINE_S do the following:
cp /sgi/local/define_s/check.fil .
define_s
The program will prompt you for various file names and options.
Alternatively, you can use a command file to provide default names of the files and options. To do this type:
cp /sgi/local/define_s/submit .
and edit the submit file to suit.
After running DEFINE_S you may look at the ASCII file output or you may look at the postscript graphical output similar to the figure above. To look at the postscript file you may type the following:
ghostview filename.ps &
The ASCII file output also has a pseudo-graphical output with more specific information.
On-line documentation for the ASCII input and output is here.
The various structural icons for the PostScript output are described below:
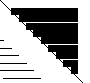 Thick lines on the diagonal indicate alpha-helix.
Thick lines on the diagonal indicate alpha-helix.
 Thin lines on the diagonal indicate beta-sheet. Intermediate lines indicate a 3-10 helix.
Thin lines on the diagonal indicate beta-sheet. Intermediate lines indicate a 3-10 helix.
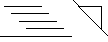 Small triangle on the diagonal indicates beta-turn.
Small triangle on the diagonal indicates beta-turn.
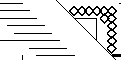 Small diamonds on the diagonal indicate omega loop.
Small diamonds on the diagonal indicate omega loop.
 Two beta-strands NOT in a beta-sheet. The axis of the left strand is shown by the vertical line in the center of the rectangle. The thick N-terminal half is at the top of the thin C-terminal half below. The axis of the lower strand is shown by the other line at the claculated interaxial angle. Again, the N-terminal half is thick, the C-terminal half thin.
Two beta-strands NOT in a beta-sheet. The axis of the left strand is shown by the vertical line in the center of the rectangle. The thick N-terminal half is at the top of the thin C-terminal half below. The axis of the lower strand is shown by the other line at the claculated interaxial angle. Again, the N-terminal half is thick, the C-terminal half thin.
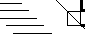 Small box on diagonal indicates a cis-proline.
Small box on diagonal indicates a cis-proline.
 A thick line box encloses helix-helix pairs which are proximal according to the following line in the file "check.fil":
A thick line box encloses helix-helix pairs which are proximal according to the following line in the file "check.fil":
15.0, CHKHHD FOR SUPERSS -max DMIN-helix-helix
The axis of the left strand is shown by the vertical line in the center of the rectangle. The thick N-terminal half is at the top of the thin C-terminal half below. The axis of the lower strand is shown by the other line at the claculated interaxial angle. Again, the N-terminal half is thick, the C-terminal half thin.
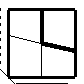 A medium line box encloses the strand-strand pairs which are proximal according to the following line in the file "check.fil":
A medium line box encloses the strand-strand pairs which are proximal according to the following line in the file "check.fil":
6.0, CHKSDT FOR BSHEET -max dist close pairs
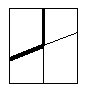 A thin line box encloses the helix-strand pairs which are proximal according to the following line in the file "check.fil":
A thin line box encloses the helix-strand pairs which are proximal according to the following line in the file "check.fil":
10.0, CHKHSD FOR SUPERSS -max DMIN-helix/strand
A similar box is present for strand-helix pairs.
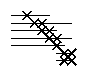 Parallel beta-sheet. The X's mark those atom pairs that are closest in a hydrogen-bonded sheet structure, either parallel or antiparallel. The lines extending to either side of the X show the range over which the distances match those expected for the strand adjacent to that atom.
Parallel beta-sheet. The X's mark those atom pairs that are closest in a hydrogen-bonded sheet structure, either parallel or antiparallel. The lines extending to either side of the X show the range over which the distances match those expected for the strand adjacent to that atom.
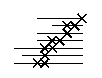 Anti beta-sheet. The X's and lines as in the paralellel beta-sheet above.
Anti beta-sheet. The X's and lines as in the paralellel beta-sheet above.
Please Note: If you publish a figure using this program please cite the following:
F.M. Richards and C.E. Kundrot (1988) Identification of Structural Motifs From Protein Coordinate Data: Secondary Structure and First-Level Supersecondary Structure. PROTEINS: Structure, Function, and Genetics 3:71-84.
Last modified: