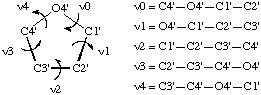|
An RNA Structure Primer |
||||||
1. What is RNA?RNA is ribonucleic acid, a close cousin of deoxyribonucleic acid or DNA. RNA is a polymer of ribonucleoside-phosphates. It's backbone is comprised of alternating ribose and phosphate groups. Ribose is a five carbon sugar that is found in a puranose, or five-membered ring, form in RNA. The phosphate groups link consecutive ribose groups and each bear one negative charge. Each monomer also has a nitrogenous base for a side chain. The four commonly found side chains in RNA are adenine, cytosine, guanine and uracil. Several other bases are occasionaly found in RNAs including: thymine, pseudouridine and methylated cytosine and guanine. Inside of cells, there are three major types of RNA: messenger RNA (mRNA), transfer RNA (tRNA) and ribosomal RNA (rRNA). There are a number of other types of RNA present in smaller quanitites as well, including small nuclear RNA (snRNA), small nucleolar RNA (snoRNA) and the 4.5S signal recognition particle (SRP) RNA. Novel species of RNA continue to be identified. |
||||||
2. What does RNA do in cells?RNA serves a multitude of roles in living cells. These include: serving as a temporary copy of genes that is used as a template for protein synthesis (mRNA), functioning as adaptor molecules that decode the genetic code (tRNA) and catalyzing the synthesis of proteins (rRNA). There is much evidence implicating RNA structure in biological regulation and catalysis. Interestingly, RNA is the only biological polymer that serves as both a catalyst (like proteins) and as information storage (like DNA). For this reason, it has be postulated RNA, or an RNA-like molecule, was the basis of life early in evolution. |
||||||
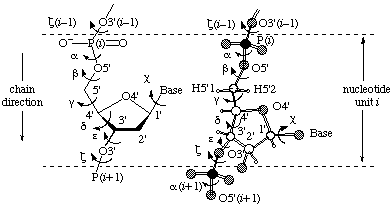
3. The Structure of RNARNA molecules are built from three basic components: ribose, a five-carbon suger, phosphate, and a family of four heterocyclic bases. The backbone of RNA is an alternating polymer of ribose and phosphate wherein phosphodiester moeities bridge the O3' and O5' atoms from consecutive riboses (Figure 1).
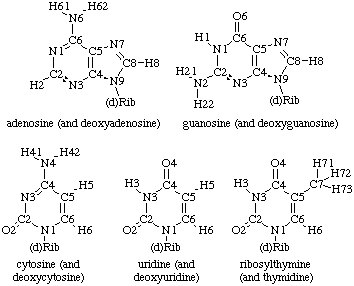
Typically, one of four heterocyclic bases is attached to the C1' atom of each ribose via a glycosidic linkage (Figure 2). These heterocycles are either purine derivatives (guanine and andenine) or pyrimidine derivatives (cytidine and uracil). Thymine, a pyrimidine normally found in DNA, is also occasionally used in RNAs. Furthermore, a large variety of base modifications are also observed in naturally occuring RNA molecules.
|
||||||
4. The Three-Dimensional Structure of RNAAlthough RNA molecules are linear polymers, they fold back on themselves to make intricate secondary and tertiary structures that are essential for them to perform their biological roles.
|
||||||
5. Describing The Three-Dimensional Structure of RNAThe three dimensional structure of molecules is usually described in Cartesian coordinates. This is the format stored in both the PDB and NDB. An equivalent description can also be given in internal coordinates. This description can be provided by given values for all the dihedral angles in a molecule. For nucleic acids, a further simplification can be made by describing the ribose ring in terms of pseudrotation. This method allows the conformation of each residue to be completely specified by five backbone dihedral angles (α, β, γ, ε and ζ) a sidechain dihedral angle (χ) and two ribose pucking paramters (P and ν0). For a definition of the backbone and sidechain dihedrals see Figure 1.
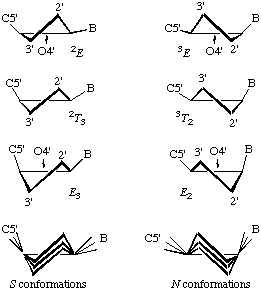
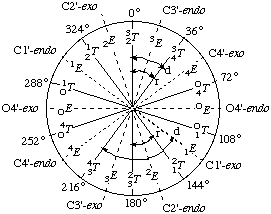
In RNABase, we use a slightly different nomenclature for the dihedral angles around the ring. We use the dihedral angles θ0, θ1, θ2, θ3, θ4. There is a simple relationship between the θ notation and the ν notation: θn = νn+2. Essentially, θ0 = ν2, θ1 = ν3, θ2 = ν4, θ3 = ν0 and θ4 = ν1. Because all of these torsion angles described rotation about bonds that are in a five-membered ring, there are relatively few combinations. Indeed, the only conformations available to ribose rings are various puckers (Figure 5).
The puckering mode can be described in terms of an amplitude (ν0), which describes the extent to which one or two atom(s) lie out of the plane of the others, and a phase (P) which describes which atom(s) lies out of the plane of the others and on which side (Figure 6).
|
||||||