Here a structure is calculated using experimentally measured interproton distance estimates, hydrogen bonds and coupling-constant-derived dihedral angle restraints. This protocol uses ab simulated annealing starting from embedded substructures using distance geometry calculations (based on the experimental data). The structure calculation is performed with the CNS task file dg_sa.inp.
cns_solve < dg_sa.inp > dg_sa.out [25 minutes]
In this protocol the extended coordinate template is used as a starting point for generation of an embedded structure. This embedded structure is generated using distance geometry calculations such that the coordinates satisfy the known geometric and experimental distrance restraints. The resulting coordinates need to be further regularized with a simulated annealing protocol. In this case five trial structures were generated. An alternative approach would have been to only generated accepted structures, in which case the script would have run until five accepted structures had been generated.
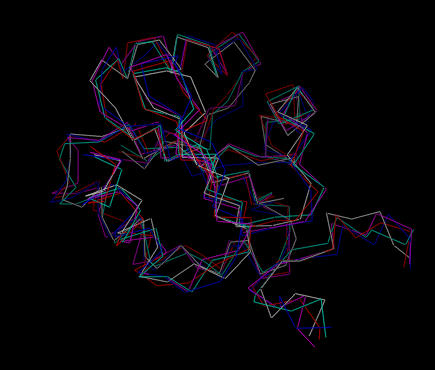
The embedded structures are written out first, prior to simulated annealing refinement. Here the names are trx_dg_sub_embed_[number].pdb. In this case, the backbone trace of the coordinates shows the overall fold of the macromolecule:
 |
| Five structures calculated by distance geometry embedding. |
After generating the embedded structures, they are further refined using simulated annealing. In this case the resulting structures have a greater variation than those generated by ab initio simulated annealing:
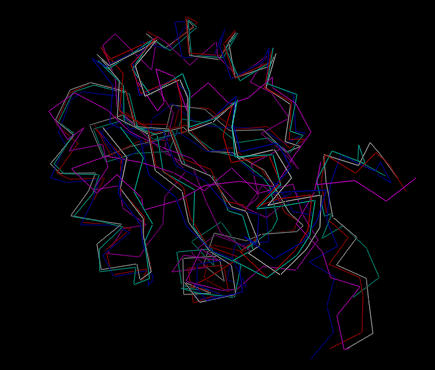 |
| Five structures after simulated annealing refinement. |
A summary of the structure calculation is written at the top of each output PDB file:
REMARK The macromolecule has 118 residues REMARK Trial structure 1 of 5 structures REMARK Molecular dynamics scheme : cartesian; cartesian; minimize REMARK High temperature dynamics : REMARK temp: 2000 steps: 1000 time(ps): 3 REMARK 1st cooling stage : REMARK temp: 2000->0 steps: 1000 time(ps): 5 temp step: 25 REMARK 2nd cooling stage not used: REMARK a total 2000 steps of minimization REMARK VDW scale factors 20 20 0.01 0.003 0.003; 3E-03->4; 1 REMARK 2617 NOEs in 2 class(es) with scale factors of 50; 50; 50 REMARK averaging function(s): sum, hbnd: sum REMARK 0 3-bond j-couplings in 0 class(es) with REMARK scale factor(s) of NA REMARK 0 1-bond j-couplings in 0 class(es) with REMARK scale factor(s) of NA REMARK 0 carbon chemical shifts in 0 class(es) with REMARK scale factor(s) of NA REMARK 0 proton chemical shifts in 0 class(es) with REMARK scale factor(s) of NA REMARK 0 diffusion anisotropy restraints in 0 class(es) with REMARK scale factor(s) of NA REMARK 0 susceptability anisotropy restraints in 0 class(es) with REMARK scale factor(s) of NA REMARK 325 dihedral restraints with scale factors of 5; 200; 400 REMARK 0 planarity restraints with a scale factor of NA REMARK NCS restraints not used. REMARK =============================================================== REMARK bond, angles, improp, vdw(<1.6), dihed REMARK violations : 2 8 1 2 41 REMARK RMSD : 0.0034 0.565 0.420 23.671 REMARK =============================================================== REMARK noe, cdih, coup, oneb, carb-a, carb-b, REMARK violations : 4 1 0 0 0 ----- REMARK RMSD : 0.030 0.513 0.000 0.000 0.000 0.000 REMARK 0.2/2 viol.: 9 5 0 REMARK =============================================================== REMARK dani, sani REMARK violations : 0 0 REMARK RMSD : 0.000 0.000 REMARK .2/.1 viol.: 0 0 REMARK =============================================================== REMARK Protons violations, rmsd REMARK all : 0 0.000 REMARK class 1: 0 0.000 REMARK class 2: 0 0.000 REMARK class 3: 0 0.000 REMARK class 4: 0 0.000 REMARK =============================================================== REMARK overall = 428.304 REMARK bon = 21.554 REMARK ang = 162.677 REMARK imp = 25.2858 REMARK vdw = 96.3987 REMARK harm = 0 REMARK noe = 111.966 REMARK coup = 0 REMARK oneb = 0 REMARK carb = 0 REMARK prot = 0 REMARK dani = 0 REMARK sani = 0 REMARK cdih = 10.4229 REMARK ncs = 0 REMARK =============================================================== REMARK DATE: 8-Jul-99 16:54:04 created by user: paul REMARK VERSION:0.5 ATOM 1 CA MET 1 10.622 -0.138 7.804 1.00 0.00 A ATOM 2 HA MET 1 10.509 -1.030 8.403 1.00 0.00 A
The information about violations can be used manually to select acceptable structures. Instead the CNS task file accept.inp can be used to identify acceptable structures and to calculate an average conformation (see previous tutorial)