cns_solve < cross_phase_phga.inp > cross_phase_phga.out [7 seconds]
Three output files are produced.
cross_phase_phga.map
cross_phase_phga_negative.peaks
cross_phase_phga_positive.peaks
The 30 highest positive and negative peaks are written in PDB format
to the *.peaks files.
If you have mapman installed, you can use the command
map_to_omap *.map
to convert the CNS maps to a format which can be read into O.
In O, enter @omac to read in the maps and
cross_phase_phga_positive.peaks.
 |
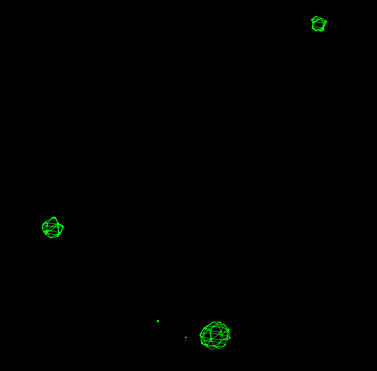
| Cross-phased map (contoured at 7.5 sigma) showing the location of mercury heavy atom sites. |
There are 3 clear peaks in the map above 7.5 sigma. These are taken as the initial sites for the mercury derivative. This is carried out by copying the cross_phase_phga_positive.peaks file to phga.pdb. This file is then edited manually to leave only the top 3 peaks and the END record:
ATOM 1 PEAK PEAK 1 14.542 9.673 43.436 1.00 23.85 PEAK ATOM 2 PEAK PEAK 2 -6.783 2.796 36.629 1.00 12.52 PEAK ATOM 3 PEAK PEAK 3 11.681 20.605 9.361 1.00 11.39 PEAK END
This PDB format file is then converted to the CNS site-database format using the CNS task file pdb_to_sdb.inp:
cns_solve < pdb_to_sdb.inp > pdb_to_sdb.out [< 1 second]
Now that we have located heavy atom sites for a uranyl and mercury derivative MIR phasing can be carried out. In this example the CNS task file mir_phase_2deriv.inp is used:
cns_solve < mir_phase_2deriv.inp > mir_phase_2deriv.inp [6 minutes]
The new MIR phases can then be used to compute other cross-phased
Fourier maps to locate sites for other derivatives, complete the heavy
atom model for the current derivatives (see next tutorial), or as
starting phases for density modification (see later tutorial).