cns_solve < fourier_phga.inp > fourier_phga.out [6 seconds]
cns_solve < gradient_phga.inp > gradient_phga.out [3.5 seconds]
Each job produces three output files.
fourier_phga.map gradient_phga.map
fourier_phga_negative.peaks gradient_phga_negative.peaks
fourier_phga_positive.peaks gradient_phga_positive.peaks
The 30 highest positive and negative peaks in both maps are
written in PDB format to the *.peaks files.
If you have mapman installed, you can use the command
map_to_omap *.map
to convert the CNS maps to a format which can be read into O.
In O, enter @omac to read in the maps and
gradient_phga_positive.peaks.
 |
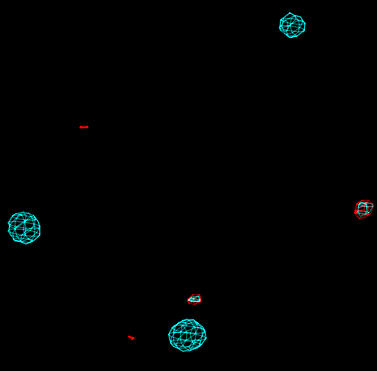
| Isomorphous difference map (cyan) and gradient map (red). Both maps are contoured at 7 sigma. |
Additional heavy-atom sites are usually characterized by peaks in both the isomorphous difference and the gradient map. In the image above, the two highest peaks in the gradient map are at locations with high densities in the isomorphous difference map. Inspection of gradient_phga_positive.peaks reveals that these are significantly greater than the other peaks:
ATOM 1 PEAK PEAK 1 25.916 22.511 26.861 1.00 13.45 PEAK ATOM 2 PEAK PEAK 2 14.713 9.913 39.363 1.00 10.43 PEAK ATOM 3 PEAK PEAK 3 -7.468 9.754 25.417 1.00 7.80 PEAK ATOM 4 PEAK PEAK 4 11.119 4.834 44.493 1.00 7.55 PEAKThese 2 top peaks are therefore added to the heavy atom model for the mercury derivative. With isomorphous replacement using heavy atoms we cannot be sure how many sites we will obtain. In particular, heavy atoms may bind weakly resulting in poorly occupied sites. It is difficult to decide at which point to ignore further peaks in the gradient and isomorphous difference maps. Fortunately, weak sites will contribute little to the phasing, and will refine to a low occupancy. Sites which are incorrectly assigned will usually refine to occupancies close to zero. Inspection of the negative peaks in the gradient map may also help indicate sites which are incorrectly placed.
The 2 top peaks from the gradient map must be added to the exisiting mercury sites. The CNS task file sdb_manipulate.inp is used to add two more entries to the phga_sites.sdb site database file:
cns_solve < sdb_manipulate.inp > sdb_manipulate.out [< 1 second]
The name of the expanded file is phga_five_sites.sdb. Use
cns_edit and copy-and-paste the coordinates from
gradient_phga_positive.peaks:
cns_edit phga_five_sites.sdb
We will assume that the same methods are used to locate further sites in the other derivative datasets. When all heavy atom models are updated then a final round of MIR phasing can be performed. In this example the CNS task file mir_phase_final.inp is used:
cns_solve < mir_phase_final.inp > mir_phase_final.out [10 minutes]
The new MIR phases can then be used to compute an electron density
map, or as starting phases for density modification (see next
tutorial).