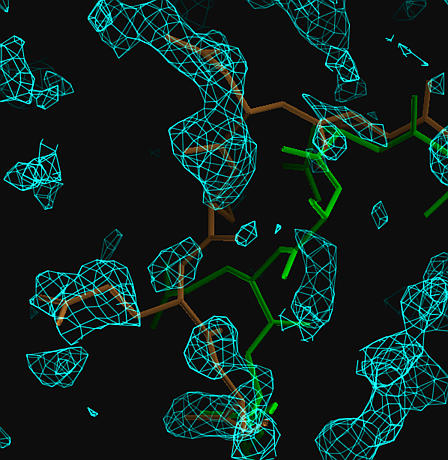
The true structure is shown in brown, the MR model in green.
Model bias is reduced around the MR coordinates.
In order to reduce the effects of model bias a simulated annealing omit map can be calculated. In the case of a molecular replacement solution we cannot be sure which parts of the model are in error therefore an omit map that covers the entire molecule is most useful. It is not possible to exclude the entire model (at most 10% can be omitted), instead small regions of the model are systematically excluded. A small map is made covering the omitted region. These small maps are accumulated and written out as a continuous map covering the whole molecule (or the defined region). Simulated annealing refinement and minimization are used to remove the bias from the omitted region. A composite omit, cross-validated, sigma-A weighted map is calculated using the CNS task file composite_omit_map.inp:
cns_solve < composite_omit_map.inp > composite_omit_map.out [10.5 hours]
The map shows less bias in regions that are incorrectlt placed.
 |
| Sigma-A weighted composite omit map (at 1.25 sigma) calculated using the MR model. The true structure is shown in brown, the MR model in green. Model bias is reduced around the MR coordinates. |