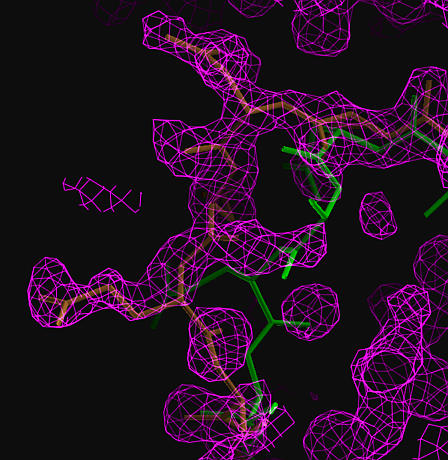
The true structure is shown in brown, the MR model in green.
The correct trace for the misplaced loop is clear.
An alternative approach to reducing model bias is to using the phases from molecular replacement as a starting point of density modification. This can be particularly powerful in cases with high non-crystallographic symmetry. In the example here we will not use the non-crystallographic symmetry because there are differences between the two monomers in the asymmetric unit. Instead density modification by solvent flipping and density truncation is used for phase improvement. First, phases and corresponding figures-of-merit must be calculated from the molecular replace model using the CNS task file model_phase.inp:
cns_solve < model_phase.inp > model_phase.out [23 seconds]
The phase probabilty distribution calculated from the model and the experimental amplitudes are used in density modifcation. Solvent flipping is used in combination with density truncation to improve phases. The mask defining the protein/solvent boundary is automatically calculated from the variation in electron density in the asymmetric unit.
cns_solve < density_modify.inp > density_modify.out [26 minutes]
The phases from density modification, the observed amplitudes and any reconstructed amplitude information (ie. unobserved data) is then used to calculate a Fourier synthesis map:
cns_solve < fourier_map.inp > fourier_map.out [23 seconds]
Density for incorrectly placed regions can be greatly improved.
 |
| FOM weighted map (at 1.5 sigma) calculated using the phases from density modification. The true structure is shown in brown, the MR model in green. The correct trace for the misplaced loop is clear. |